How to reproduce the analysis
Kevin Cazelles and Kevin Solarik
2019-10-20
Source:vignettes/Solarik.Rmd
Solarik.RmdThis vignette aims at guiding the reader through the different steps of the analysis carried out in Priority effects will prevent range shifts of temperate tree species into the boreal forest.1.
Data
Data are currently archived on Dryad at the following URL https://doi.org/10.5061/dryad.q573n5tdx and can be download using retrieveData() as follows
This create a folder (by default it is input) with all datasets as .csv or .xlsx files. Once done, trees locations and regeneration files can readily be formatted with formatData(). Note that, as favuorability data are strored as .docx files,in order to use the code below it requires to convert the .docx into .csv files.
dat_abitibi <- formatData("input/abitibitrees.csv", "input/abitibiregen.csv",
"input/abitibisubstrate.csv")
dat_lebic <- formatData("input/lebictrees.csv", "input/lebicregen.csv",
"input/lebicsubstrate.csv", abbr_site = "bic")
dat_sutton <- formatData("input/suttontrees.csv", "input/suttonregen.csv", "input/suttonsubstrate.csv", abbr_site = "sut")By default all the files created will be exported as .rds file in output. Note that we stored all these data as .rda files in data (see, for instance, ?dat_abitibi).
Sites of the study
As explained in the original study, recruitment data have been measured on three different sites:
- ‘abi’: Abitibi
- ‘bic’: Le Bic
- ‘sut’: Sutton
The map below shows the location of those sites. For each site, extra information are available on click.
Tree species of the study
| abbr1 | abbr2 | Scientific Names | TSN | abi | bic | sut |
|---|---|---|---|---|---|---|
| BF | ABBA | Abies balsamea | 18032 | TRUE | TRUE | TRUE |
| RM | ACRU | Acer rubrum | 28728 | TRUE | TRUE | FALSE |
| SM | ACSA | Acer saccharum | 28731 | TRUE | TRUE | TRUE |
| YB | BEAL | Betula alleghaniensis | 19481 | FALSE | FALSE | TRUE |
| WB | BEPA | Betula papyrifera | 19489 | TRUE | TRUE | TRUE |
| AB | FAGR | Fagus grandifolia | 19462 | FALSE | FALSE | TRUE |
| AS | POTR | Populus tremuloides | 195773 | TRUE | TRUE | TRUE |
Statistical models
Below are detailed the steps to reproduce our analysis.
Models
General equation
For a given species and a given year, the seeds recruitment depends on:
- the fecundity of surrounding individuals of the same species;
- the substrate favourability;
- the neighborhood effect.
\[R_i = \text{STR}\sum_{j=1}^Sc_{i,j}f_{i,j}\sum_{k=1}^T\left(\frac{\text{DBH}_k}{30}\right)^2 \frac{1}{K}e^{-\text{DM}_{i,k}}e^{-\frac{h_i}{1-h_i}\sum_{l=1}^Ba_{i,l}}\]
Simulations
Overall strategy
For each model, we estimate the parameters through the maximization of the likelihood (actually the minimization of the likelihood log-transformed) for the corresponding data. Every model is a combination :
- the site and the tree species considered;
- the age of seedling taken into account (1 or 2);
- whether we used a zero-inflated poisson or a regular poisson distribution;
- the kernel dispersal used;
- the distribution used to model seedling recruitment (zero-inflated poisson or a regular poisson).
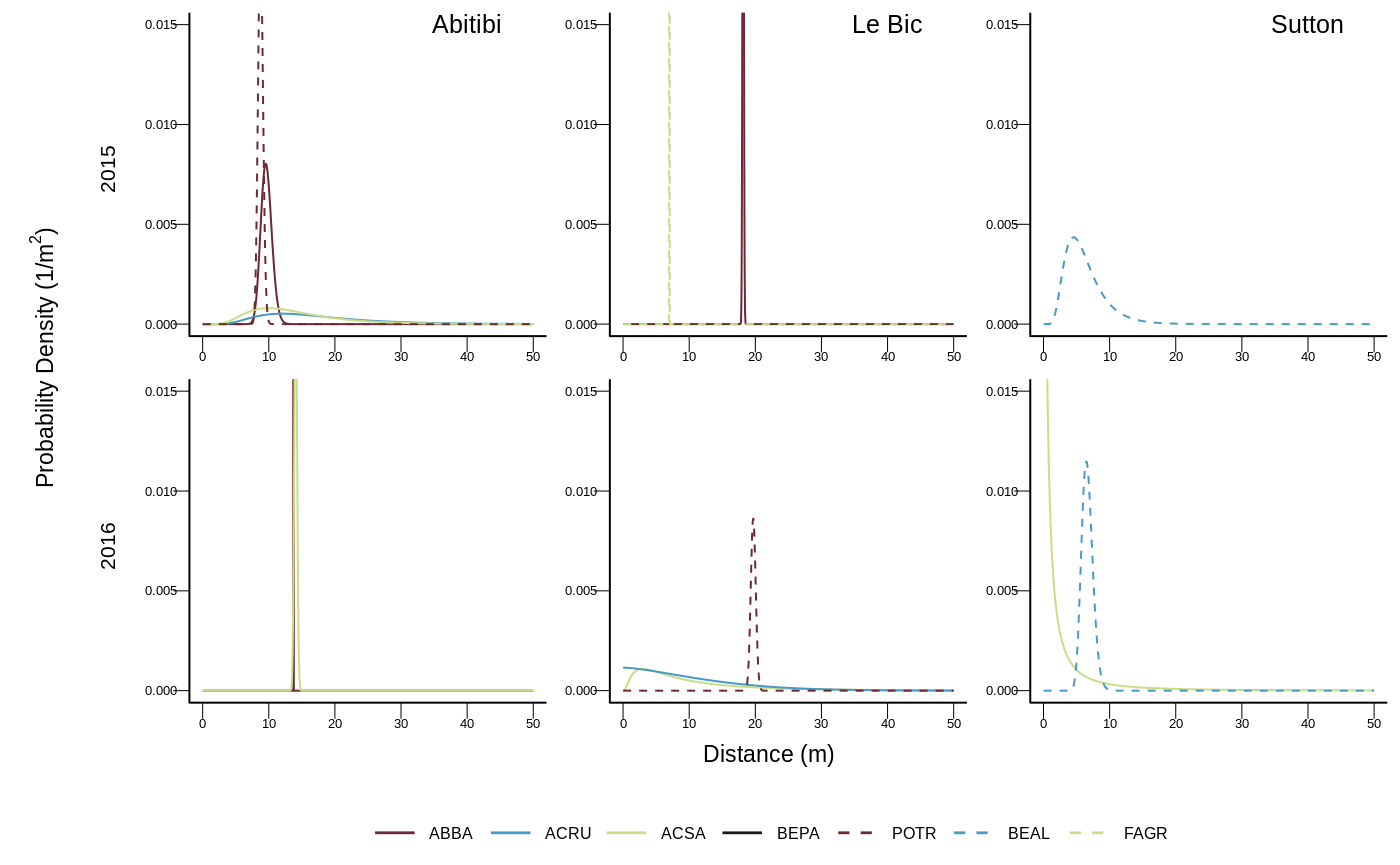
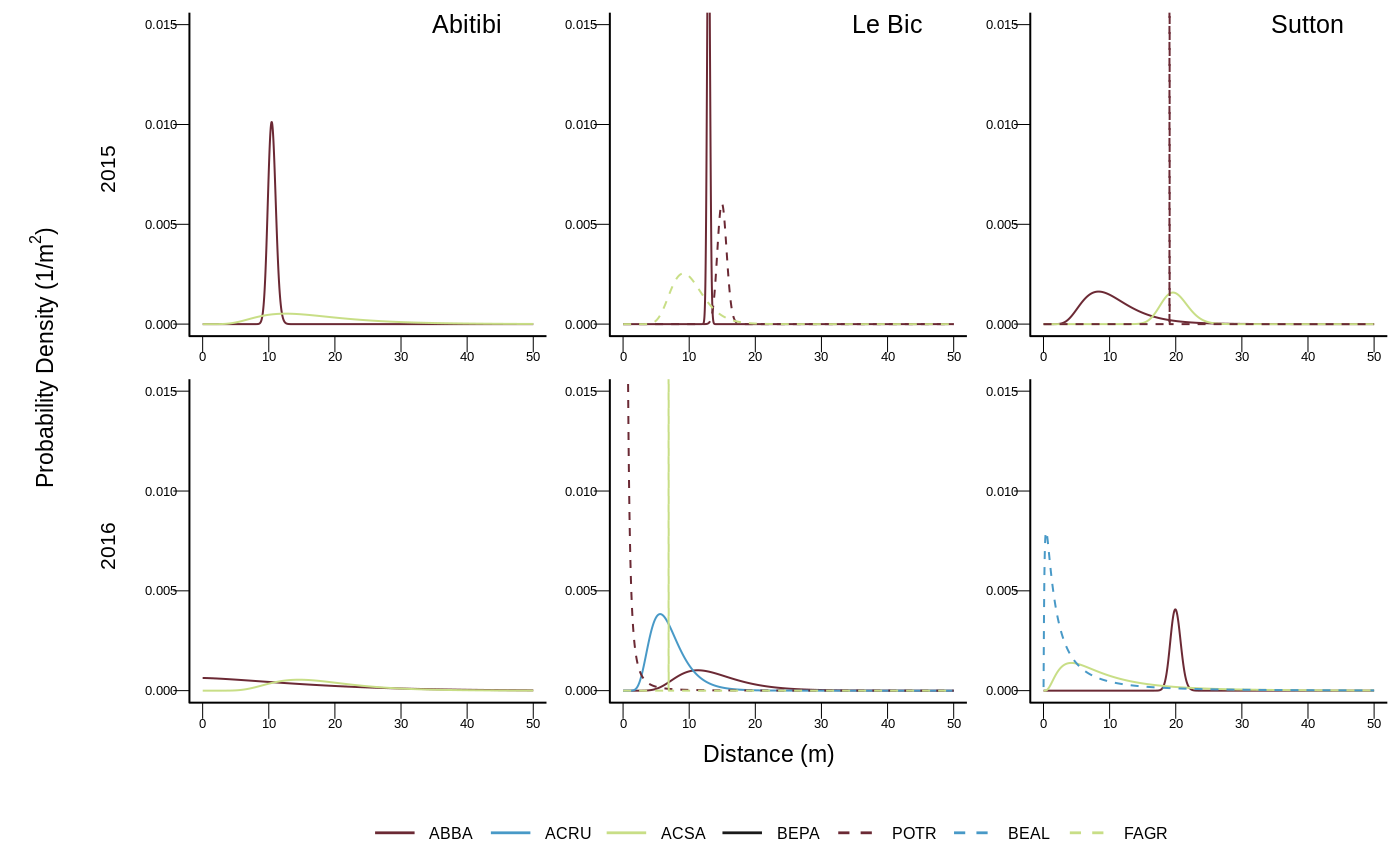
As an example, below are the 15 first models out of the 320 tested for Abies balsamea:
| site | tree | year | age | pz | disp | favo | neigh |
|---|---|---|---|---|---|---|---|
| abi | ABBA | 2015 | 1 | TRUE | 0 | TRUE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 1 | TRUE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 2 | TRUE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 3 | TRUE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 4 | TRUE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 0 | FALSE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 1 | FALSE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 2 | FALSE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 3 | FALSE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 4 | FALSE | TRUE |
| abi | ABBA | 2015 | 1 | TRUE | 0 | TRUE | FALSE |
| abi | ABBA | 2015 | 1 | TRUE | 1 | TRUE | FALSE |
| abi | ABBA | 2015 | 1 | TRUE | 2 | TRUE | FALSE |
| abi | ABBA | 2015 | 1 | TRUE | 3 | TRUE | FALSE |
| abi | ABBA | 2015 | 1 | TRUE | 4 | TRUE | FALSE |
As the models we used are not linear and highly customized we optimized the likelihood with the simulated annealing algorithm implemented in GenSA2.
Numerical implementation
For each row of simuDesign, columns are arguments passed to recruitmentAnalysis() in which:
parameters are computed for the simulated annealing (
getParameters());simulated annealing is performed (
GenSA()from GenSA package) to minimize the likelihood (getLikelihood()).
| STR | Pz | scal | shap | pm | pl | pn | pd | ps | pb | |
|---|---|---|---|---|---|---|---|---|---|---|
| start | 1e+02 | 0.5 | 1.00 | 2 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 1 |
| low | 0e+00 | 0.0 | 0.02 | 1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0 |
| high | 1e+06 | 1.0 | 20.00 | 4 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1000 |
Example
The line below (not run) allows to run for the 16th model.
res <- launchIt(iter = 16, simu = simuDesign, mxt = 100,
record = "output/test.txt", quiet = TRUE, path = "output/")
# value (likelihood)
res$value
# parameters value
res$parNote that test.txt records all the values tested during the simulated algorithm and the corresponding likelihood based on which we obtain a confident interval.
Solarik et al. DOI: 2Badded↩
Y. Xiang, S. Gubian. B. Suomela, J. Hoeng (2013). Generalized Simulated Annealing for Efficient Global Optimization: the GenSA Package for R. The R Journal, Volume 5/1, June 2013. https://journal.r-project.org/archive/2013/RJ-2013-002/index.html↩